Like my previous post, this blog is also motivated by a comment by Professor Persi Diaconis in his recent Stanford probability seminar. The seminar was about a way of “collapsing” a random walk on a group to a random walk on the set of double cosets. In this post, I’ll first define double cosets and then go over the example Professor Diaconis used to make us probabilists and statisticians more comfortable with all the group theory he was discussing.
Double cosets
Let be a group and let
and
be two subgroups of
. For each
, the
-double coset containing
is defined to be the set
To simplify notation, we will simply write double coset instead of -double coset. The double coset of
can also be defined as the equivalence class of
under the relation
for some
and
Like regular cosets, the above relation is indeed an equivalence relation. Thus, the group can be written as a disjoint union of double cosets. The set of all double cosets of
is denoted by
. That is,
Note that if we take , the trivial subgroup, then the double cosets are simply the left cosets of
,
. Likewise if
, then the double cosets are the right cosets of
,
. Thus, double cosets generalise both left and right cosets.
Double cosets in 
Fix a natural number . A partition of
is a finite sequence
such that
,
and
. For each partition of
,
, we can form a subgroup
of the symmetric group
. The subgroup
contains all permutations
such that
fixes the sets
. Meaning that
for all
. Thus, a permutation
must individually permute the elements of
, the elements of
and so on. This means that, in a natural way,
If we have two partitions and
, then we can form two subgroups
and
and consider the double cosets
. The claim made in the seminar was that the double cosets are in one-to-one correspondence with
contingency tables with row sums equal to
and column sums equal to
. Before we explain this correspondence and properly define contingency tables, let’s first consider the cases when either
or
is the trivial subgroup.
Left cosets in 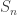
Note that if , then
is the trivial subgroup and, as noted above,
is simply equal to
. We will see that the cosets in
can be described by forgetting something about the permutations in
.
We can think of the permutations in as all the ways of drawing without replacement
balls labelled
. We can think of the partition
as a colouring of the
balls by
colours. We colour balls
by the first colour
, then we colour
the second colour
and so on until we colour
the final colour
. Below is an example when
is equal to 6 and
.

Note that a permutation is in
if and only if we draw the balls by colour groups, i.e. we first draw all the balls with colour
, then we draw all the balls with colour
and so on. Thus, continuing with the previous example, the permutation
below is in
but
is not in
.

It turns out that we can think of the cosets in as what happens when we “forget” the labels
and only remember the colours of the balls. By “forgetting” the labels we mean only paying attention to the list of colours. That is for all
,
if and only if the list of colours from the draw
is the same as the list of colours from the draw
. Thus, the below two permutations define the same coset of

To see why this is true, note that if and only if
for some
. Furthermore,
if and only if
maps each colour group to itself. Recall that function composition is read right to left. Thus, the equation
means that if we first relabel the balls according to
and then draw the balls according to
, then we get the result as just drawing by
. That is,
for some
if and only if drawing by
is the same as first relabelling the balls within each colour group and then drawing the balls according to
. Thus,
, if and only if when we forget the labels of the balls and only look at the colours,
and
give the same list of colours. This is illustrated with our running example below.

Right cosets of 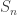
Typically, the subgroup is not a normal subgroup of
. This means that the right coset
will not equal the left coset
. Thus, colouring the balls and forgetting the labelling won’t describe the right cosets
. We’ll see that a different type of forgetting can be used to describe
.
Fix a partition and now, instead of considering
colours, think of
different people
. As before, a permutation
can be thought of drawing
balls labelled
without replacement. We can imagine giving the first
balls drawn to person
, then giving the next
balls to the person
and so on until we give the last
balls to person
. An example with
and
is drawn below.

Note that if and only if person
receives the balls with labels
in any order. Thus, in the below example
but
.

It turns out the cosets are exactly determined by “forgetting” the order in which each person
received their balls and only remembering which balls they received. Thus, the two permutation below belong to the same coset in
.

To see why this is true in general, consider two permutations . The permutations
result in each person
receiving the same balls if and only if after
we can apply a permutation
that fixes each subset
and get
. That is,
and
result in each person
receiving the same balls if and only if
for some
. Thus,
are the same after forgetting the order in which each person received their balls if and only if
. This is illustrated below,

We can thus see why . A left coset
correspond to pre-composing
with elements of
and a right cosets
correspond to post-composing
with elements of
.
Contingency tables
With the last two sections under our belts, describing the double cosets is straight forward. We simply have to combine our two types of forgetting. That is, we first colour the
balls with
colours according to
. We then draw the balls without replace and give the balls to
different people according
. We then forget both the original labels and the order in which each person received their balls. That is, we only remember the number of balls of each colour each person receives. Describing the double cosets by “double forgetting” is illustrated below with
and
.

The proof that double forgetting does indeed describe the double cosets is simply a combination of the two arguments given above. After double forgetting, the number of balls given to each person can be recorded in an table. The
entry of the table is simply the number of balls person
receives of colour
. Two permutations are the same after double forgetting if and only if they produce the same table. For example,
and
above both produce the following table
| Green ( | Red ( | Blue ( | Total | |
| Person | 1 | 0 | 1 | 2 |
| Person | 1 | 1 | 0 | 2 |
| Person | 1 | 1 | 0 | 2 |
| Total | 3 | 2 | 1 | 6 |
By the definition of how the balls are coloured and distributed to each person we must have for all and
and
An table with entries
satisfying the above conditions is called a contingency table. Given such a contingency table with entries
where the rows sum to
and the columns sum to
, there always exists at least one permutation
such that
is the number of balls received by person
of colour
. We have already seen that two permutations produce the same table if and only if they are in the same double coset. Thus, the double cosets
are in one-to-one correspondence with such contingency tables.
The hypergeometric distribution
I would like to end this blog post with a little bit of probability and relate the contingency tables above to the hyper geometric distribution. If for some
, then the contingency tables described above have two rows and are determined by the values
in the first row. The numbers
are the number of balls of colour
the first person receives. Since the balls are drawn without replacement, this means that if we put the uniform distribution on
, then the vector
follows the multivariate hypergeometric distribution. Thus, if we have a random walk on
that quickly converges to the uniform distribution on
, then we could use the double cosets to get a random walk that converges to the multivariate hypergeometric distribution (although there are smarter ways to do such sampling).